|
Size: 266
Comment:
|
← Revision 21 as of 2014-12-02 18:35:27 ⇥
Size: 1901
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| --------- ------- |
|
| Line 3: | Line 5: |
| ||||'''''Antonella Succurro, PhD'''''|| ||<|2> {{attachment:profilepic.png}} ||[[attachment:resume_antonella.pdf|download CV]]|| ||in [[http://www.accliphot.eu/|AccliPhot]] since September 8, 2014|| |
||||<tablewidth="573px" tableheight="303px"style="border:none; ;text-align:center">'''''Antonella Succurro, PhD''''' || ||||<style="border:none; ;text-align:center"> || ||<style="border:none; ;text-align:center" |7> {{attachment:profilepic.png|New Orleans, August 2014|width="200"}} ||<style="border:none;"> || ||<style="border:none; ;text-align:center">in [[http://www.accliphot.eu/|AccliPhot]] since September 8, 2014 || ||<style="border:none; ;text-align:center">''Institute for Quantitative and Theoretical Biology'' || ||<style="border:none; ;text-align:center">Heinrich Heine Universität Düsseldorf || ||<style="border:none; ;text-align:right">[[attachment:resume_antonella.pdf|download CV]] || ||<style="border:none; ;text-align:right">[[http://personal.ifae.es/succurro/thesis/succurro_phdthesis_web.pdf|my PhD thesis]] || ||<style="border:none; ;text-align:right"> a.succurro [AT] gmail.com|| ------- --------- === Main Project === '''''Mathematical Modelling of Chloroplast Signalling Pathways''''' |
| Line 8: | Line 23: |
| === Main Project === | === Secondary Project === {{http://www.bu.edu/segrelab/files/2013/07/newbiomass.gif|COMETS at work|align="right", width=500}} '''''Simulation of algae and bacteria consortium''''' During a short-term visiting position at Boston University, I collaborate with Daniel Segrè's team towards the implementation of algae model in the COMETS framework. [[http://www.bu.edu/segrelab/comets/|COMETS (Computation of Microbial Ecosystems in Time and Space)]] is a software developed at Daniel Segrè Lab to simulate with dynamic flux balance analysis genome-scale metabolic networks of microbial colonies diffusing and interacting on a 2D space lattice. [[https://git.hhu.de/succurro/algae-bacteria/wikis/home]][[Project's wiki page]] (accessible only to collaborators) |
Antonella Succurro, PhD |
|
|
|
|
|
in AccliPhot since September 8, 2014 |
|
Institute for Quantitative and Theoretical Biology |
|
Heinrich Heine Universität Düsseldorf |
|
a.succurro [AT] gmail.com |
|
Main Project
Mathematical Modelling of Chloroplast Signalling Pathways
Secondary Project
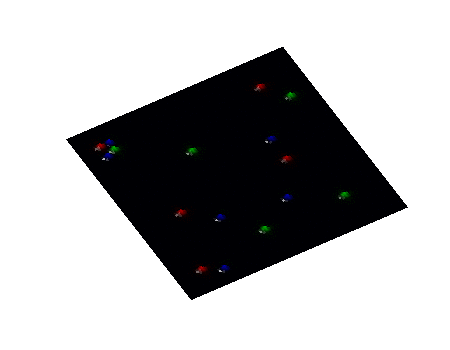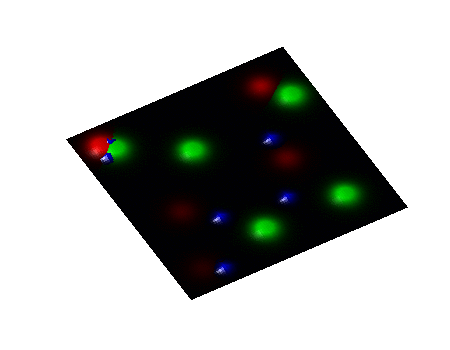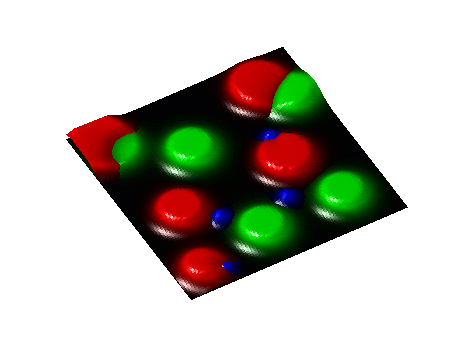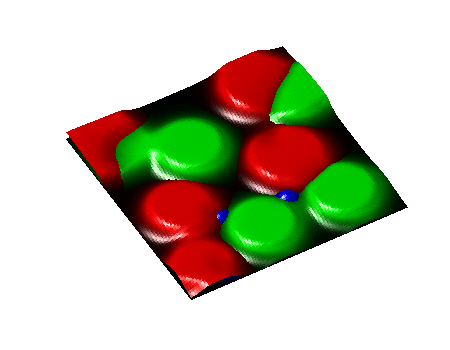 Simulation of algae and bacteria consortium
Simulation of algae and bacteria consortium
During a short-term visiting position at Boston University, I collaborate with Daniel Segrè's team towards the implementation of algae model in the COMETS framework. COMETS (Computation of Microbial Ecosystems in Time and Space) is a software developed at Daniel Segrè Lab to simulate with dynamic flux balance analysis genome-scale metabolic networks of microbial colonies diffusing and interacting on a 2D space lattice.
https://git.hhu.de/succurro/algae-bacteria/wikis/homeProject's wiki page (accessible only to collaborators)
