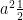|
Size: 1885
Comment:
|
Size: 1923
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 5: | Line 5: |
| Our group began nearly thirty years ago with initial interests in computer simulation of metabolism and the theoretical analysis of metabolic control and regulation. Whilst these still remain areas of interest, we have since developed interests in modelling signal transduction, in various different approaches to network analysis of metabolism, and in reconstructing metabolic networks from genomic data. In the course of this research, we have addressed problems in microbial, plant and mammalian metabolism, often in conjunction with collaborators who have contributed experimental results. | Our '''group''' began nearly thirty years ago with initial interests in computer simulation of metabolism and the theoretical analysis of metabolic control and regulation. Whilst these still remain areas of interest, we have since developed interests in modelling signal transduction, in various different approaches to network analysis of metabolism, and in reconstructing metabolic networks from genomic data. In the course of this research, we have addressed problems in microbial, plant and mammalian metabolism, often in conjunction with collaborators who have contributed experimental results. |
| Line 10: | Line 10: |
<<latex($a^2 \frac{1}{2}$)>> |
News:
Advance notice. The Biochemical Society has approved plans for the 71st Harden Conference 19-23 September 2011: Metabolic Pathway Analysis 3.
Our group began nearly thirty years ago with initial interests in computer simulation of metabolism and the theoretical analysis of metabolic control and regulation. Whilst these still remain areas of interest, we have since developed interests in modelling signal transduction, in various different approaches to network analysis of metabolism, and in reconstructing metabolic networks from genomic data. In the course of this research, we have addressed problems in microbial, plant and mammalian metabolism, often in conjunction with collaborators who have contributed experimental results.
Our current work centres on modelling the networks of reactions in cells, with particular emphasis on metabolism. It forms part of the emerging field of Systems Biology, in that we are concerned with understanding how biological function arises from the interactions between many components, and with building predictive models. We have to develop and apply suitable theoretical tools, including metabolic control analysis, computer simulation and other forms of algebraic and numerical analysis. In addition, we are investigating how to decipher the metabolic information contained in genome sequences. We are involved in projects on microbial, plant and animal metabolism, each in collaboration with an experimental team.
Potential applications of our work include the design of changes in cellular metabolism to improve the output of product such as antibiotics, detecting vulnerable sites in cellular networks that could be targets for drugs to control disease-causing organisms, and improved understanding of how organisms manage to adjust their metabolism in response to environmental changes and other signals.