|
Size: 246
Comment:
|
Size: 1774
Comment: First go at instructions.
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 5: | Line 5: |
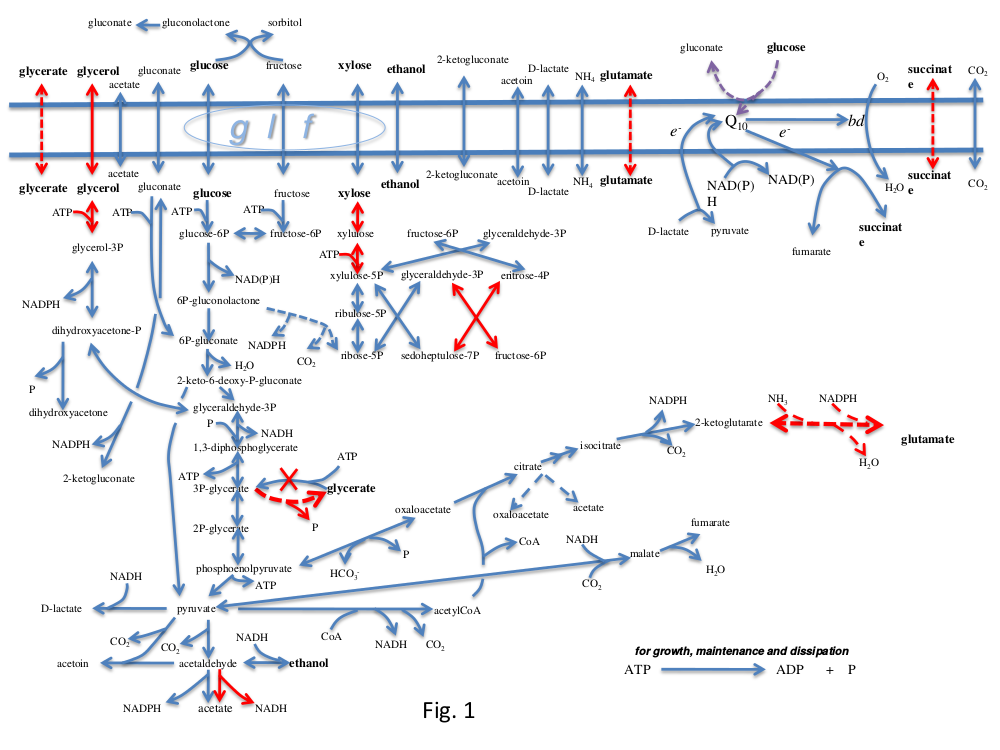
| Groups 1, 3 and 5 should start with this exercise | Groups 1, 3 and 5 should start with this exercise. Your job is to engineer ''Z. mobilis'' to try and make ethanol from xylose, which is derived from the hemicellulose in lignocellulosic biomass. 1. Into your model from the previous exercise, you need to extend the ''Include'' directive to load a file of additional reactions from ''Z. mobilis'' metabolism contained in the file ZmCHO.spy. These reactions will incorporate a number of the essential reactions marked with blue arrows in the diagram below. Check the elementary modes of this extended model. Does it produce any products from xylose? 1. Create a new text file "Heterologous.spy" into which you can write (in !ScrumPy syntax) the reactions of the enzymes you intend to engineer ''Z. mobilis'' to express. Again, this file will be linked into the model via the ''Include'' directive. 1. Reactions marked in red in the diagram are not present natively in the organism, so you will need to add a selection of these to enable ethanol formation from xylose. Find the EC numbers of the enzymes you need. The instructions in the previous part of the practical show you how to get the !MetaCyc reactions vis !PyoCyc. Copy the needed reactions from the !ScrumPy window into the new file. 1. Reload your modified model. Check whether additional vectors appear in the null space. Also check for Orphan metabolites. Whenthe model looks as though it may be working, compute the elementary modes. 1. Have you got modes from xylose to ethanol? What is the carbon conversion efficiency? |
Practical 5: group work
Exercise 1
Groups 1, 3 and 5 should start with this exercise.
Your job is to engineer Z. mobilis to try and make ethanol from xylose, which is derived from the hemicellulose in lignocellulosic biomass.
Into your model from the previous exercise, you need to extend the Include directive to load a file of additional reactions from Z. mobilis metabolism contained in the file ZmCHO.spy. These reactions will incorporate a number of the essential reactions marked with blue arrows in the diagram below. Check the elementary modes of this extended model. Does it produce any products from xylose?
Create a new text file "Heterologous.spy" into which you can write (in ScrumPy syntax) the reactions of the enzymes you intend to engineer Z. mobilis to express. Again, this file will be linked into the model via the Include directive.
Reactions marked in red in the diagram are not present natively in the organism, so you will need to add a selection of these to enable ethanol formation from xylose. Find the EC numbers of the enzymes you need. The instructions in the previous part of the practical show you how to get the MetaCyc reactions vis PyoCyc. Copy the needed reactions from the ScrumPy window into the new file.
- Reload your modified model. Check whether additional vectors appear in the null space. Also check for Orphan metabolites. Whenthe model looks as though it may be working, compute the elementary modes.
- Have you got modes from xylose to ethanol? What is the carbon conversion efficiency?
Exercise 2
Groups 2 and 4 should start with this exercise
Zymomonas metabolism