|
Size: 4154
Comment: Linked in groups list
|
← Revision 10 as of 2014-12-18 15:18:18 ⇥
Size: 5423
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
== Late addition (sorry!) == The transaldolase reaction went missing from ZmCHO.spy. Either download it again, or else manually add the reaction: {{{ # EC-2.2.1.2 "TRANSALDOL-RXN": "GAP" + "D-SEDOHEPTULOSE-7-P"<>"FRUCTOSE-6P" + "ERYTHROSE-4P" ~ #REVERSIBLE }}} |
|
| Line 21: | Line 34: |
| 1. If you've got time left after this exercise, go to Exercise 2. | === Solution === If from "Zymomonas.spy" you included "ZmED.spy" (with corrections, but minimal reactions commented out), "ZmCHO.spy" and [[http://mudsharkstatic.brookes.ac.uk/Download/Models/C1Net/HeterologousX.spy|HeterologousX.spy]], you should get an elementary mode from xylose to ethanol, CO_2 and acetate. The acetate formation is inevitable, and the pathway involves cycling pentose through the pentose phosphate reactions. |
| Line 40: | Line 54: |
| 1. Reload your modified model. Check whether additional vectors appear in the null space. Also check for Orphan metabolites. Bear in mind that reactions in the original !ZmEd.spy file that weren't needed to make ethanol from glucose might be necessary on the path from xylose. When the model looks as though it may be working, compute the elementary modes. | 1. Reload your modified model. Check whether additional vectors appear in the null space. Also check for Orphan metabolites. Bear in mind that reactions in the original !ZmEd.spy file that weren't needed to make ethanol from glucose might be necessary on the path from glycerol. When the model looks as though it may be working, compute the elementary modes. |
| Line 42: | Line 56: |
| 1. Have you got modes from xylose to ethanol? What is the carbon conversion efficiency? | 1. Have you got modes from glycerol to ethanol? If not, is there a redox problem. ''Z. mobilis'' has an electron transport chain hat is largely uncoupled from phosphorylation. Let us represent this as: {{{ "OxPhos": "NADH" + x_O2 -> "NAD" + "WATER" ~ }}} 1. Does this help? If so, What is the carbon conversion efficiency? |
| Line 44: | Line 64: |
| 1. If you've got time left after this exercise, go to Exercise 2. | === Solution === If from "Zymomonas.spy" you included "ZmED.spy" (with corrections, but minimal reactions commented out), "ZmCHO.spy" and [[http://mudsharkstatic.brookes.ac.uk/Download/Models/C1Net/HeterologousG.spy|HeterologousG.spy]], you should get an elementary mode from glycerol to ethanol and CO_2, but only one ethanol per glycerol, and oxidation is needed as glycerol is more reduced than glucose. |
Practical 5: group work
First find out which group you are in and join your fellow members.
Late addition (sorry!)
- The transaldolase reaction went missing from ZmCHO.spy. Either download it again, or else manually add the reaction:
# EC-2.2.1.2 "TRANSALDOL-RXN": "GAP" + "D-SEDOHEPTULOSE-7-P"<>"FRUCTOSE-6P" + "ERYTHROSE-4P" ~ #REVERSIBLE
Exercise 1
Groups 1, 3 and 5 should start with this exercise.
Your job is to engineer Z. mobilis to try and make ethanol from xylose, which is derived from the hemicellulose in lignocellulosic biomass.
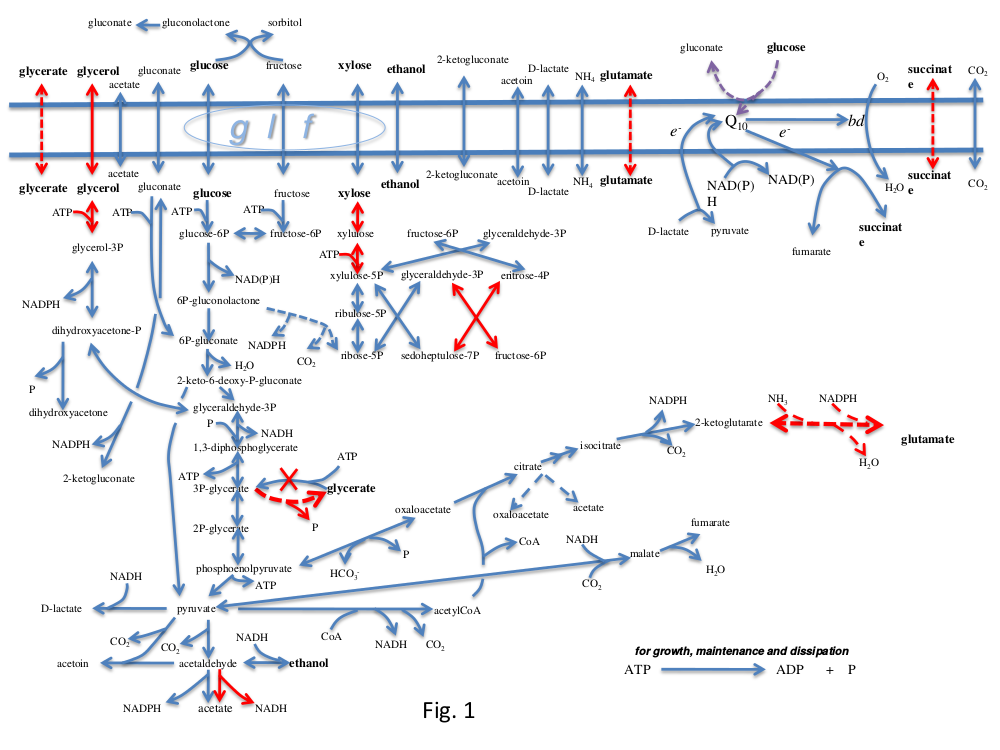
Into your model from the previous exercise, you need to extend the Include directive to load a file of additional reactions from Z. mobilis metabolism contained in the file ZmCHO.spy. These reactions will incorporate a number of the essential reactions marked with blue arrows in the diagram below. Check the elementary modes of this extended model. Does it produce any products from xylose?
Create a new text file "Heterologous.spy" into which you can write (in ScrumPy syntax) the reactions of the enzymes you intend to engineer Z. mobilis to express. Again, this file will be linked into the model via the Include directive.
Reactions marked in red in the diagram are not present natively in the organism, so you will need to add a selection of these to enable ethanol formation from xylose. Find the EC numbers of the enzymes you need. The instructions in the previous part of the practical show you how to get the MetaCyc reactions vis PyoCyc. Copy the needed reactions from the ScrumPy window into the new file.
Reload your modified model. Check whether additional vectors appear in the null space. Also check for Orphan metabolites. Bear in mind that reactions in the original ZmEd.spy file that weren't needed to make ethanol from glucose might be necessary on the path from xylose. When the model looks as though it may be working, compute the elementary modes.
- Have you got modes from xylose to ethanol? What is the carbon conversion efficiency?
Solution
If from "Zymomonas.spy" you included "ZmED.spy" (with corrections, but minimal reactions commented out), "ZmCHO.spy" and HeterologousX.spy, you should get an elementary mode from xylose to ethanol, CO_2 and acetate. The acetate formation is inevitable, and the pathway involves cycling pentose through the pentose phosphate reactions.
Exercise 2
Groups 2 and 4 should start with this exercise.
Your job is to engineer Z. mobilis to try and make ethanol from glycerol, which is cheaply available from the manufacture of biodiesel.
Into your model from the previous exercise, you need to extend the Include directive to load a file of additional reactions from Z. mobilis metabolism contained in the file ZmCHO.spy. These reactions will incorporate a number of the essential reactions marked with blue arrows in the diagram below. Check the elementary modes of this extended model to make sure it has some working metabolism. It does not at the moment use glycerol.
Create a new text file "Heterologous.spy" into which you can write (in ScrumPy syntax) the reactions of the enzymes you intend to engineer Z. mobilis to express. Again, this file will be linked into the model via the Include directive. The first reaction you need to add is a glycerol transporter:
Glycerol_tx: x_Glycerol <> "GLYCEROL" ~
Reactions marked in red in the diagram are not present natively in the organism, so you will need to add a selection of these to enable ethanol formation from glycerol. Find the EC numbers of the enzymes you need. The instructions in the previous part of the practical show you how to get the MetaCyc reactions vis PyoCyc. Copy the needed reactions from the ScrumPy window into the new file.
Reload your modified model. Check whether additional vectors appear in the null space. Also check for Orphan metabolites. Bear in mind that reactions in the original ZmEd.spy file that weren't needed to make ethanol from glucose might be necessary on the path from glycerol. When the model looks as though it may be working, compute the elementary modes.
Have you got modes from glycerol to ethanol? If not, is there a redox problem. Z. mobilis has an electron transport chain hat is largely uncoupled from phosphorylation. Let us represent this as:
"OxPhos": "NADH" + x_O2 -> "NAD" + "WATER" ~
- Does this help? If so, What is the carbon conversion efficiency?
Solution
If from "Zymomonas.spy" you included "ZmED.spy" (with corrections, but minimal reactions commented out), "ZmCHO.spy" and HeterologousG.spy, you should get an elementary mode from glycerol to ethanol and CO_2, but only one ethanol per glycerol, and oxidation is needed as glycerol is more reduced than glucose.
Zymomonas metabolism