|
Size: 6080
Comment:
|
Size: 6633
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
| The general form of the description of a kinetic model has been covered in the pages on the [[../../../SpyMDL|ScrumPy Model Description Language]]. Consider this [[http://mudsharkstatic.brookes.ac.uk/Delhi2013/Models/TwoReacRever.spy | model file]] an example. It | The general form of the description of a kinetic model has been covered in the pages on the [[../../../SpyMDL|ScrumPy Model Description Language]]. Consider this [[http://mudsharkstatic.brookes.ac.uk/Delhi2013/Models/TwoReacRever.spy | model file]] an example. It |
| Line 9: | Line 9: |
| There are two limitations on the syntax for kinetic models: 1. The identifiers cannot be quoted, and 1. No c or C++ key words can be used as identifiers. |
There are two limitations on the syntax for kinetic models: 1. The identifiers cannot be quoted, and 1. No c or C++ key words can be used as identifiers. |
| Line 13: | Line 13: |
| On the other hand, it is possible to compute the values to be assigned, as in this example of adjusting an equilibrium constant to its apparent value at the prevailing pH: {{{ |
On the other hand, it is possible to compute the values to be assigned, as in this example of adjusting an equilibrium constant to its apparent value at the prevailing pH: {{{ |
| Line 19: | Line 19: |
| If the two reaction model above is saved as a ''spy'' file, it can be loaded into !ScrumPy to create an instance of a model object as follows: {{{ |
If the two reaction model above is saved as a ''spy'' file, it can be loaded into !ScrumPy to create an instance of a model object as follows: {{{ |
| Line 25: | Line 25: |
| Once it has been loaded, values can be inspected, e.g.: {{{ |
Once it has been loaded, values can be inspected, e.g.: {{{ |
| Line 35: | Line 35: |
| They can, of course, also be changed: {{{ |
They can, of course, also be changed: {{{ |
| Line 41: | Line 41: |
| However, note: {{{ |
However, note: {{{ |
| Line 49: | Line 49: |
| That is, the computation of the rate is not updated until ''m.Eval()'' is called to force a re-evaluation of the rate with current parameters and concentration values. | That is, the computation of the rate is not updated until ''m.Eval()'' is called to force a re-evaluation of the rate with current parameters and concentration values. |
| Line 51: | Line 51: |
| The following table shows the methods/attributes of any model instance ''m'' that return lists of the corresponding entities: || '''Constituent''' || '''Values''' || '''Identifiers''' || || Parameters || m.!GetVec(!ScrumPy.Param) || m.!ParamNames || || Metabolites || m.!GetVec(!ScrumPy.Conc) || m.sm.rnames || || Metabolite rates of change || m.!GetVec(!ScrumPy.dMet) || m.sm.rnames || || Reaction rates || m.!GetVec(!ScrumPy.Vel) || m.sm.cnames || The corresponding method ''!SetVec'' takes as parameters one of the same codes as !GetVec followed by a vector of values, but the vector must be the correct length ( as indicated by, e.g. ''len(m.!ParamNames)''). |
The following table shows the methods/attributes of any model instance ''m'' that return lists of the corresponding entities: || '''Constituent''' || '''Values''' || '''Identifiers''' || || Parameters || m.!GetVec(!ScrumPy.Param) || m.!ParamNames || || Metabolites || m.!GetVec(!ScrumPy.Conc) || m.sm.rnames || || Metabolite rates of change || m.!GetVec(!ScrumPy.dMet) || m.sm.rnames || || Reaction rates || m.!GetVec(!ScrumPy.Vel) || m.sm.cnames || The corresponding method ''!SetVec'' takes as parameters one of the same codes as !GetVec followed by a vector of values, but the vector must be the correct length ( as indicated by, e.g. ''len(m.!ParamNames)''). |
| Line 61: | Line 61: |
| ScrumPy takes care of any dependencies between metabolites by computing the moiety conservation relationships from the stoichiometry matrix. These arise, for example, where coenzymes such as NAD and NADH occur in a model, but the nucleotide moieties are not synthesised and degraded by the model reactions. | ScrumPy takes care of any dependencies between metabolites by computing the moiety conservation relationships from the stoichiometry matrix. These arise, for example, where coenzymes such as NAD and NADH occur in a model, but the nucleotide moieties are not synthesised and degraded by the model reactions. |
| Line 63: | Line 63: |
| In such cases, a new parameter, with an identifier starting ''CSUM_'', is created and assigned a value that is the sum of the initialised concentrations of the metabolites in the conservation group. One of the metabolite concentrations will then not be integrated, but will have its concentration determined as the difference between the CSUM parameter and the sum of all the remaining metabolite concentrations. | In such cases, a new parameter, with an identifier starting ''CSUM_'', is created and assigned a value that is the sum of the initialised concentrations of the metabolites in the conservation group. One of the metabolite concentrations will then not be integrated, but will have its concentration determined as the difference between the CSUM parameter and the sum of all the remaining metabolite concentrations. |
| Line 65: | Line 65: |
| It is strongly recommended that you do not try to anticipate dependencies by manually eliminating one of the conserved group metabolites from your model. ScrumPy uses a mathematically-validated method to determine the conservation relationships, which can be complicated and non-intuitive, especially where two conservation groups intersect. Furthermore, without the conservation sum constraint, steady state solvers cannot be guaranteed to respect the conserved total. | It is strongly recommended that you do not try to anticipate dependencies by manually eliminating one of the conserved group metabolites from your model. ScrumPy uses a mathematically-validated method to determine the conservation relationships, which can be complicated and non-intuitive, especially where two conservation groups intersect. Furthermore, without the conservation sum constraint, steady state solvers cannot be guaranteed to respect the conserved total. |
| Line 67: | Line 67: |
| As an example, if the model [[http://mudsharkstatic.brookes.ac.uk/Delhi2013/Models/SimpleCycle.spy | SimpleCycle.spy]] is downloaded, saved and load into !ScrumPy, the following commands can be issued: {{{ |
As an example, if the model [[http://mudsharkstatic.brookes.ac.uk/Delhi2013/Models/SimpleCycle.spy | SimpleCycle.spy]] is downloaded, saved and load into !ScrumPy, the following commands can be issued: {{{ |
| Line 83: | Line 83: |
| ''m.!ParamNames'' shows that an additional papameter, ''CSUM_NADH'' has been created, and ''m.!GetVec(!ScrumPy.Param)'' shows this has been initialised as 1. ''m.!ConsMoieties()'' returns a single vector showing there is one conservation relationship, involving 1 NAD and 1 NADH. (1/1 is the rational fraction representation of 1). ''m.!ConsDic()'' conveys the same information, but returns the dependency as a dictionary, with ''mpq(1,1)'' being another representation of the rational fraction 1/1. | ''m.!ParamNames'' shows that an additional papameter, ''CSUM_NADH'' has been created, and ''m.!GetVec(!ScrumPy.Param)'' shows this has been initialised as 1. ''m.!ConsMoieties()'' returns a single vector showing there is one conservation relationship, involving 1 NAD and 1 NADH. (1/1 is the rational fraction representation of 1). ''m.!ConsDic()'' conveys the same information, but returns the dependency as a dictionary, with ''mpq(1,1)'' being another representation of the rational fraction 1/1. |
| Line 88: | Line 88: |
| The steady state of the model can be determined with ''m.FindSS()''; if the steady state is found successfully, than ''m.IsOK()'' will return ''True''. | The steady state of the model can be determined with ''m.FindSS()''; if the steady state is found successfully, than ''m.IsOK()'' will return ''True''. |
| Line 90: | Line 90: |
| A straightforward way to record a series of steady states as a parameter is changed uses the '''AddStatMonitor''' method. For example: {{{ |
A straightforward way to record a series of steady states as a parameter is changed uses the '''AddStatMonitor''' method. For example: {{{ |
| Line 108: | Line 108: |
| resulting in | resulting in |
| Line 110: | Line 110: |
| {{attachment:PlotSSeg.png}} | {{attachment:PlotSSeg.png}} |
| Line 114: | Line 114: |
| Before looking for potential problems with the numerical solution of your model, check via ''structural analysis'' (insert links) that your model has elementary modes connecting inputs to outputs, and that there are no structurally dead reactions (other than those that can attain equilibrium). | Before looking for potential problems with the numerical solution of your model, check via ''structural analysis'' (insert links) that your model has elementary modes connecting inputs to outputs, and that there are no structurally dead reactions (other than those that can attain equilibrium). |
| Line 116: | Line 116: |
| Options in FindSS and pre-simulation. | The ''FindSS'' method may not converge if the initial estimates of the variables are not close enough to the steady-state values. One way to move the variables closer to their steady state values is by a time course simulation. In fact, the ''FindSS'' method will automatically try this if it initially fails to find a steady state. However, the simulation time it uses is relatively short. Hence it might be worth explicitly running a time course simulation (see LINK) to see whether the model is or is not appearing to tend to a stable steady state. |
Kinetic Modelling
Contents
1. Loading a model
The general form of the description of a kinetic model has been covered in the pages on the ScrumPy Model Description Language. Consider this model file an example. It
contains two reaction definitions with their kinetic equations, followed by initial assignments of the parameters and the variable (B).
- There are two limitations on the syntax for kinetic models:
- The identifiers cannot be quoted, and
- No c or C++ key words can be used as identifiers.
pH = 7.5 k2eq = 17.84e8*10**(-pH)
If the two reaction model above is saved as a spy file, it can be loaded into ScrumPy to create an instance of a model object as follows:
m = ScrumPy.Model()
2. Inspecting and assigning values
- Once it has been loaded, values can be inspected, e.g.:
>>> m[ " x_A " ] # parameter 1 . 0 >>> m[ "B" ] # concentration 0.4 >>> m[ "R1" ] # rate, evaluated with current variable values 3.3333333333333335
>>> m[ " x_A " ] = 2 # parameter >>> m[ "B" ] = 2 # concentration
>>> m["R1"] 3.3333333333333335 >>> m.Eval() >>> m["R1"] 2.0
That is, the computation of the rate is not updated until m.Eval() is called to force a re-evaluation of the rate with current parameters and concentration values.
The following table shows the methods/attributes of any model instance m that return lists of the corresponding entities:
Constituent
Values
Identifiers
Parameters
m.GetVec(ScrumPy.Param)
m.ParamNames
Metabolites
m.GetVec(ScrumPy.Conc)
m.sm.rnames
Metabolite rates of change
m.GetVec(ScrumPy.dMet)
m.sm.rnames
Reaction rates
m.GetVec(ScrumPy.Vel)
m.sm.cnames
The corresponding method SetVec takes as parameters one of the same codes as GetVec followed by a vector of values, but the vector must be the correct length ( as indicated by, e.g. len(m.ParamNames)).
2.1. Moiety conservation parameters
ScrumPy takes care of any dependencies between metabolites by computing the moiety conservation relationships from the stoichiometry matrix. These arise, for example, where coenzymes such as NAD and NADH occur in a model, but the nucleotide moieties are not synthesised and degraded by the model reactions.
In such cases, a new parameter, with an identifier starting CSUM_, is created and assigned a value that is the sum of the initialised concentrations of the metabolites in the conservation group. One of the metabolite concentrations will then not be integrated, but will have its concentration determined as the difference between the CSUM parameter and the sum of all the remaining metabolite concentrations.
It is strongly recommended that you do not try to anticipate dependencies by manually eliminating one of the conserved group metabolites from your model. ScrumPy uses a mathematically-validated method to determine the conservation relationships, which can be complicated and non-intuitive, especially where two conservation groups intersect. Furthermore, without the conservation sum constraint, steady state solvers cannot be guaranteed to respect the conserved total.
As an example, if the model SimpleCycle.spy is downloaded, saved and load into ScrumPy, the following commands can be issued:
>>> m.ParamNames ['x_A', 'x_C', 'CSUM_NADH'] >>> m.GetVec(ScrumPy.Param) [2.0, 1.0, 1.0] >>> m.ConsMoieties() NAD NAD 1/1 B 0/1 C 0/1 NADH 1/1 >>> m.ConsDic() {'NAD': [('NADH', mpq(1,1))]}
m.ParamNames shows that an additional papameter, CSUM_NADH has been created, and m.GetVec(ScrumPy.Param) shows this has been initialised as 1. m.ConsMoieties() returns a single vector showing there is one conservation relationship, involving 1 NAD and 1 NADH. (1/1 is the rational fraction representation of 1). m.ConsDic() conveys the same information, but returns the dependency as a dictionary, with mpq(1,1) being another representation of the rational fraction 1/1.
3. Examining steady states of the model
The steady state of the model can be determined with m.FindSS(); if the steady state is found successfully, than m.IsOK() will return True.
A straightforward way to record a series of steady states as a parameter is changed uses the AddStatMonitor method. For example:
>>> results=m.AddStatMonitor() >>> m["x_A"]=1 >>> m.FindSS() >>> for n in range(20): m["Vmax1"] += 1 m.FindSS() print m.IsOK()
results is a ScrumPy matrix, the columns of which (results.cnames) contain the time, rates, concentrations and parameter values of the model (though the time is always zero in this case). A row is added to the matrix with each successful steady state determination, recording the steady state values.
There are methods for plotting the outcome of the set of analyses that are described more fully in Secondary Analysis of Model Results (link to be made). A simple example is:
>>> results.SetPlotX("Vmax1") >>> results.AddToPlot("R1")resulting in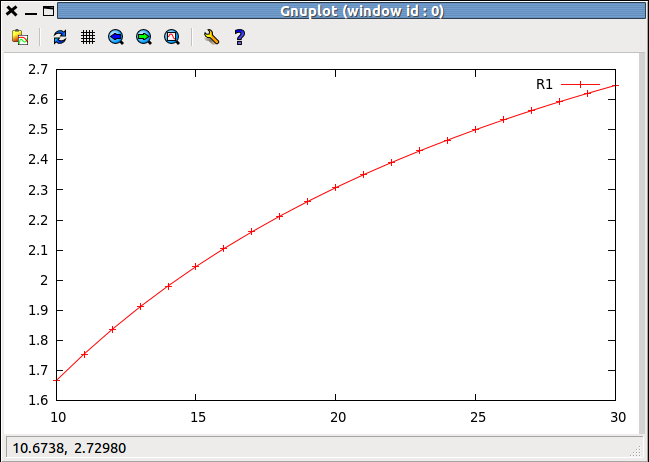
3.1. What to do if a steady state is not found
Before looking for potential problems with the numerical solution of your model, check via structural analysis (insert links) that your model has elementary modes connecting inputs to outputs, and that there are no structurally dead reactions (other than those that can attain equilibrium).
The FindSS method may not converge if the initial estimates of the variables are not close enough to the steady-state values. One way to move the variables closer to their steady state values is by a time course simulation. In fact, the FindSS method will automatically try this if it initially fails to find a steady state. However, the simulation time it uses is relatively short. Hence it might be worth explicitly running a time course simulation (see LINK) to see whether the model is or is not appearing to tend to a stable steady state.